What causes genetic drift in populations?
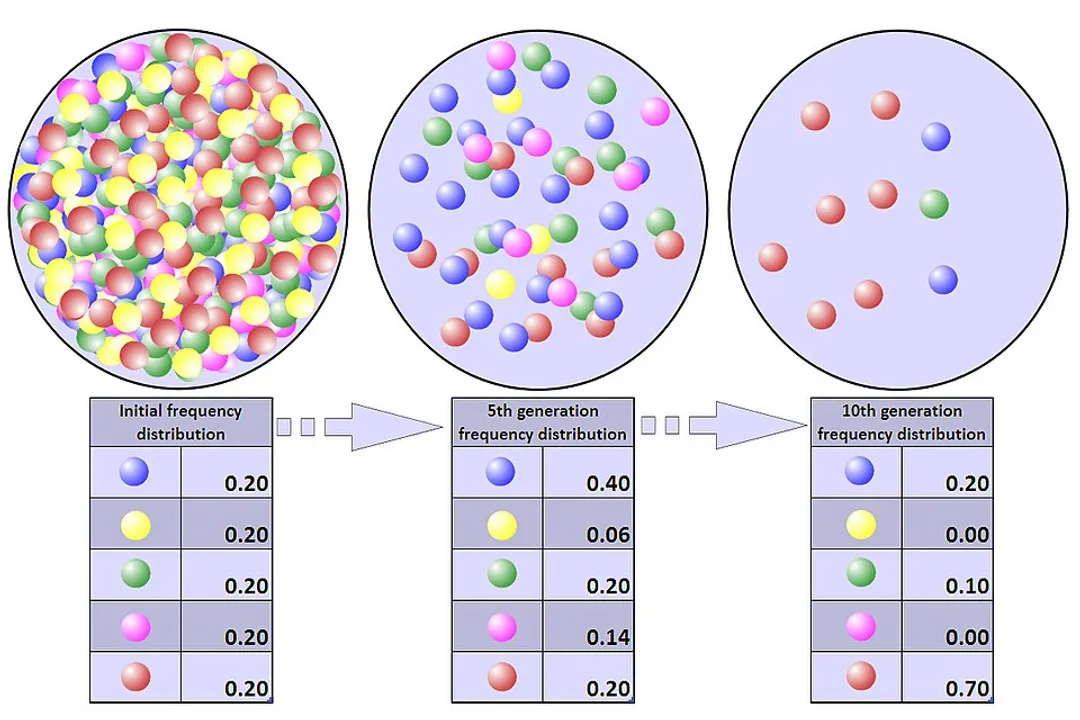
Genetic drift describes a fundamental mechanism of evolution where allele frequencies in a population change from one generation to the next due to chance events. [2][5] Unlike natural selection, which is driven by fitness advantages, genetic drift is entirely random, meaning that the alleles that increase or decrease in frequency are not related to whether they are helpful or harmful to the organism's survival or reproduction. [5][7] It is essentially a form of sampling error occurring across generations. [1]
Imagine flipping a coin ten times. You expect five heads and five tails, but you might easily get seven heads and three tails just by luck. If those coin flips represented which individuals in a tiny population successfully passed on their genes, the resulting gene pool would no longer be perfectly 50/50, simply because of the random nature of who happened to be sampled into the next generation. [3][7] This randomness means drift can cause evolution to occur even in the absence of any selective pressure. [5]
# Random Sampling
The entire process of genetic drift hinges on the fact that a finite sample of genes—the survivors and reproducers—is used to build the next generation. [7] Not every individual in the parent generation will reproduce, and even among those that do, the exact combination of alleles passed on is subject to chance. [1]
Consider a very small, isolated population of flowering plants where the gene for flower color has two alleles: red (R) and white (r). If, by random chance, a strong wind or a localized patch of disease happens to wipe out most of the plants carrying the recessive white (r) allele before they can flower and disperse seeds, the next generation will likely have a much higher frequency of the red (R) allele, not because red flowers were "better," but because the white-flowered individuals were unlucky. [1][5]
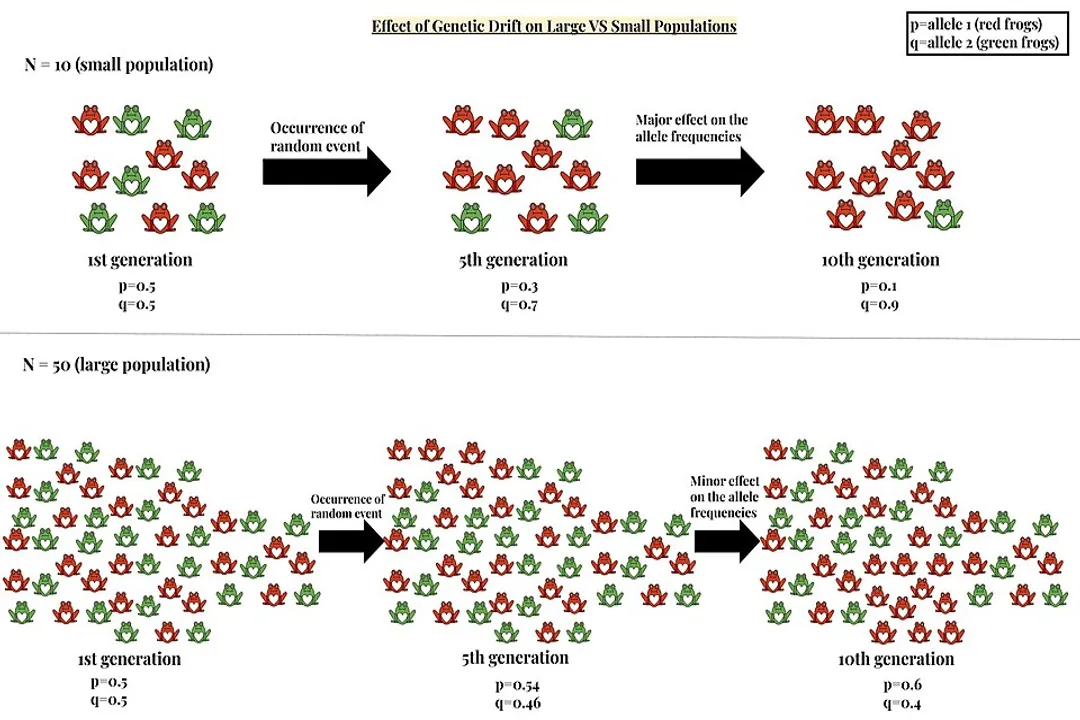
The magnitude of this effect is inversely proportional to the size of the population. [7] In very large populations, the random sampling error is usually negligible because the sheer number of individuals averages out most minor fluctuations. If you flip a coin a million times, you will get very close to a 50/50 split. [5] In contrast, if the population is small, a single chance event can dramatically shift the genetic makeup of the entire group, driving the change much further and faster. [7][9]
If we look at the mathematical relationship, the variance in allele frequency change due to drift is proportional to , where is the population size. [7] This simple formula encapsulates why drift is a far more potent evolutionary force in small groups. Think about a small, isolated island community founded by just twenty people. If three of those twenty people happen to carry a rare, recessive disorder allele, that allele starts with a representation in the gene pool. If a larger continental population of twenty million people only has a few dozen carriers of that same allele, the impact of those few individuals being randomly excluded from reproduction is virtually zero compared to the island group. [1]
# Population Size
The concept of population size is central to understanding the causes and strength of genetic drift. While we often talk about the census size—the actual number of individuals—evolutionary biologists pay close attention to the effective population size (). [9] The is a theoretical number that accounts for factors that reduce the actual reproductive contribution of the population, such as unequal sex ratios, differences in reproductive success among individuals, or fluctuating population sizes over time. [9] A population might have 1,000 members counted, but if only 50 females successfully breed each year while the males vary widely in success, the effective size might be much closer to 100, meaning drift effects will be magnified as if the population were only 100 strong. [9]
When genetic drift operates strongly in small populations, it leads to rapid consequences:
- Loss of Heterozygosity: Genetic variation decreases because alleles are randomly lost from the gene pool. [7]
- Fixation: An allele can eventually reach a frequency of 100% (fixation), meaning every individual in the population has that allele, or it can be lost entirely (frequency of 0%). [7] Once an allele is fixed or lost through drift, the only way to reintroduce variation at that specific locus is through new mutation or gene flow from another population. [7]
It is worth noting that while drift is random regarding which alleles change frequency, it is non-random regarding which alleles are most likely to be affected: alleles with small initial frequencies are much more susceptible to complete loss than common alleles. [5] This erosion of rare alleles can sometimes remove beneficial variants before selection ever has a chance to act upon them. [5]
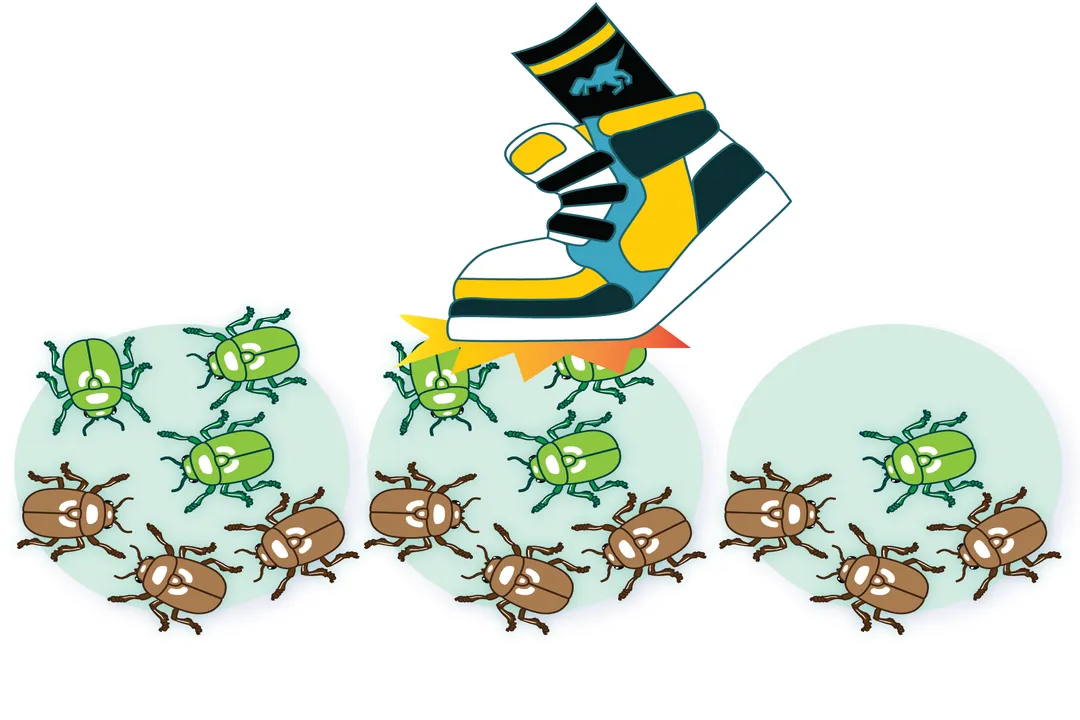
# Bottleneck Events
One major scenario that dramatically causes genetic drift to become the dominant evolutionary force is the population bottleneck. [1] This occurs when a population's size is severely reduced for at least one generation due to some external, random environmental event. [1]
These events are indiscriminate; they kill individuals based on chance, not fitness. Examples include natural disasters like floods, fires, earthquakes, or sudden, widespread disease outbreaks. [1] Because the surviving group is a small, random sample of the original, much larger gene pool, the resulting population will likely have allele frequencies that differ significantly from the ancestral population. [1][8]
Imagine a large, diverse population of wild sheep. A harsh, unexpected winter followed by a spring avalanche wipes out 95% of the flock. The few dozen survivors, carrying only a subset of the original genetic diversity, form the basis for the population's future recovery. [1] If the original population had a slightly higher frequency of an allele conferring resistance to a future parasite, but that allele happened to be concentrated by chance in the individuals killed by the avalanche, the recovering population will now be poorly equipped to deal with that parasite, even though the environment didn't change its selective pressure. [8]
To illustrate this, consider a hypothetical population of 1,000 beetles where 50% are green and 50% are brown. If a quick, unselective pesticide application kills 950 beetles, leaving 50 survivors, it is highly improbable that the survivors will still be exactly 50% green and 50% brown. They might randomly end up being 70% brown and 30% green. This shift in frequency—the cause of the drift—is immediate and powerful in the subsequent generations rebuilding from that small base. [1]
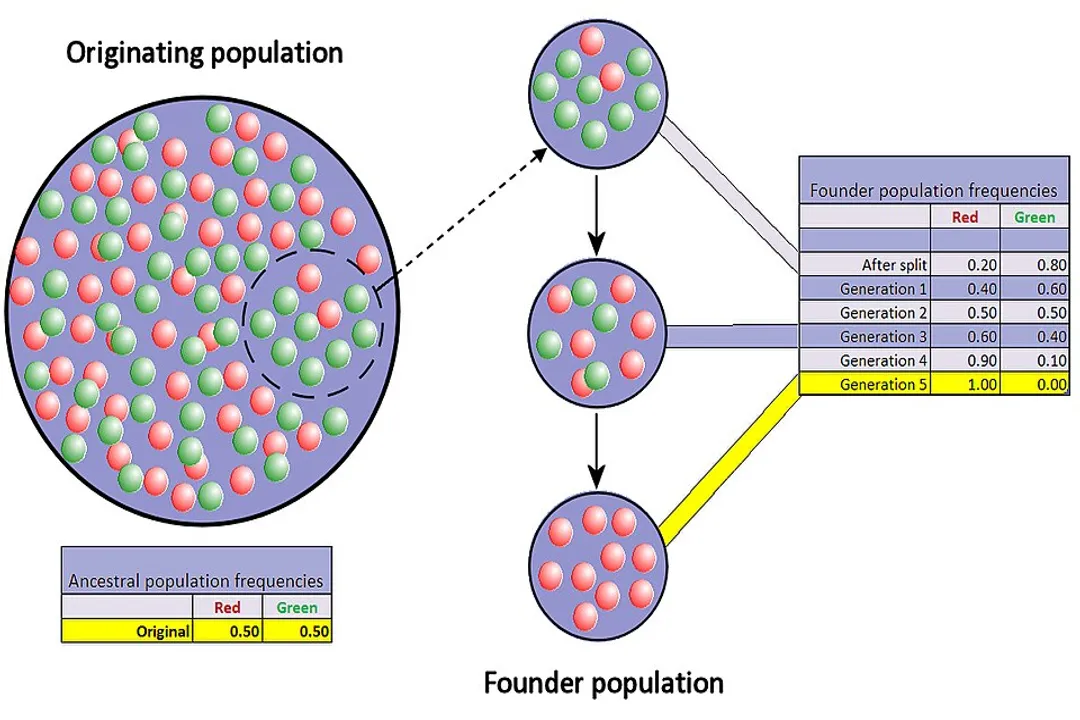
# Founder Effect
The Founder Effect is another critical cause of rapid, strong genetic drift, often observed in colonization events. [1] This happens when a small group breaks away from a larger parent population to establish a new, isolated colony. [1]
The genetic makeup of this founding group is unlikely to perfectly represent the diversity of the original source population simply because only a few individuals are available to migrate. [1] The new population’s entire gene pool is derived from those few founders.
This phenomenon is frequently seen in island biogeography or in human populations that have expanded from a small number of initial migrants. [1] For example, if a small group of birds is blown off course to an isolated island, the alleles they happen to carry—including any rare or deleterious ones—will immediately become the dominant genetic material for that new population. [1] If, by random chance, one of the founders happened to carry a specific, rare marker allele, that allele will be much more common in the island population than it ever was on the mainland, purely due to its initial representation in the founding sample. [8]
This is different from a bottleneck because the population never existed at a large size in that location; the drift occurs at the moment of establishment due to the small initial sample size. A population starting with only thirty individuals experiences the same intensity of random fluctuation as a population of thirty survivors from a disaster.
# Contextual Analysis
It is easy to view genetic drift as purely a historical or natural process, but its modern causes are often linked to human activity that fragments landscapes. When large continuous habitats are broken up into smaller, isolated patches—a process known as habitat fragmentation—we are, in essence, creating dozens of small, unrelated populations where only one large one existed before. [9] Each of these small patches then becomes subject to intense, localized genetic drift. Unlike the large ancestral population where selection pressures could act consistently across millions of individuals, these fragmented groups suffer rapid loss of heterozygosity independently. This means that even if the species as a whole is resilient, the individual sub-populations within those fragments may become genetically impoverished and less able to adapt to future, specific environmental changes within their patch. [9] We are engineering the conditions for drift to accelerate across the species range.
Furthermore, it is important to recognize that selection and drift are not always mutually exclusive; they often act simultaneously, though one may dominate depending on the context. [5] In large populations, selection is the primary driver of predictable change. However, drift can introduce significant noise into that process. If an allele provides a fitness advantage of just 1%, selection will slowly favor it. But if the population is small, genetic drift might randomly wipe out that slightly beneficial allele before selection can build up its frequency. This competitive interaction means that effective population size becomes a critical determinant of whether an advantageous mutation successfully spreads or is lost due to the luck of the draw. [5][9]
# Conclusion
Genetic drift is caused by the inescapable reality of random chance affecting gene transmission across generations, particularly when the population size is small enough that sampling error becomes significant. [2][7] Whether initiated by a sudden catastrophe leading to a bottleneck, or by the isolation of a few colonists establishing a founder effect, the result is the same: a non-adaptive shift in the genetic structure of the descendants. [1] Understanding drift requires appreciating the population size—the effective size, —as the primary moderator of its power, constantly eroding the raw material of evolution, genetic diversity, through sheer statistical bad luck. [5][9]
#Videos
Genetic Drift - YouTube
Related Questions
#Citations
Genetic drift (article) | Natural selection - Khan Academy
Genetic Drift - National Human Genome Research Institute
Genetic Drift - YouTube
How does genetic drift work? : r/evolution - Reddit
Genetic drift - Understanding Evolution
Genetic Drift - an overview | ScienceDirect Topics
Genetic drift - Wikipedia
Genetic Drift - American Phytopathological Society
Genetic Drift and Effective Population Size | Learn Science at Scitable